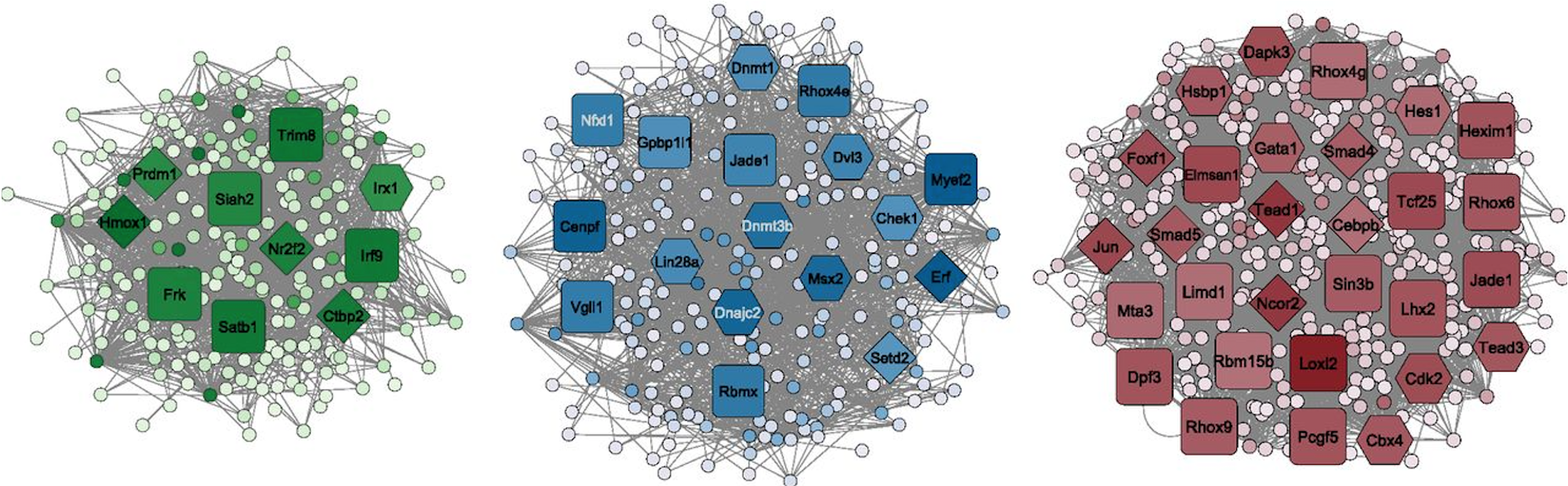
An article from the Tuteja lab, "Identifying novel regulators of placental development using time-series transcriptome data" was published in Life Science Alliance today! Ha Vu, a Bioinformatics and Computational Biology graduate student, is lead author on the manuscript. She analyzed RNA-seq data we generated from mouse fetal placenta tissues at embryonic day (e) 7.5, e8.5 and e9.5 to identify genes with timepoint-specific expression, then inferred gene interaction networks to analyze highly connected network modules. We determined that timepoint-specific gene network modules associated with distinct developmental processes, and with similar expression profiles to specific human placental cell populations. From each module, we obtained hub genes and their direct neighboring genes, which were predicted to govern placental functions. Haninder Kaur in our lab confirmed that four novel candidate regulators identified through our analyses regulate cell migration in vitro. In summary, we were able to predict several novel regulators of placental development using network analysis of bulk RNA-seq data. Our findings and analysis approaches could be used to study other tissues, or for future studies investigating the transcriptional landscape of early placental development.
Identifying novel regulators of placental development using time-series transcriptome data12/13/2022
0 Comments
Your comment will be posted after it is approved.
Leave a Reply. |